Data Engineering - Week 2
Week 2 - Data Engineering Zoomcamp course: Data ingestion
- Data Lake
- Data Lake vs Data Warehouse
- ETL vs ELT
- Workflow Orchestration
- Airflow
- Transfer Service in GCP
- Conclusion
Note: The content of this post is from the course videos, my understandings and searches, and reference documentations.
Data Lake
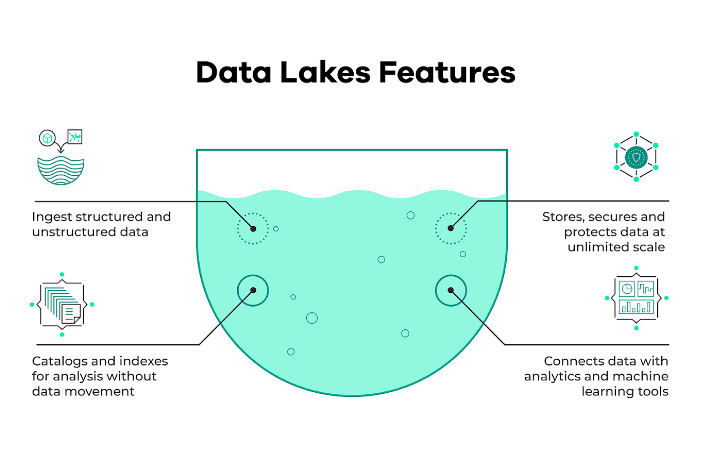
A data lake is a collection of technologies that enables querying of data contained in files or blob objects. When used effectively, they enable massive scale and cost-effective analysis of structured and unstructured data assets [source].
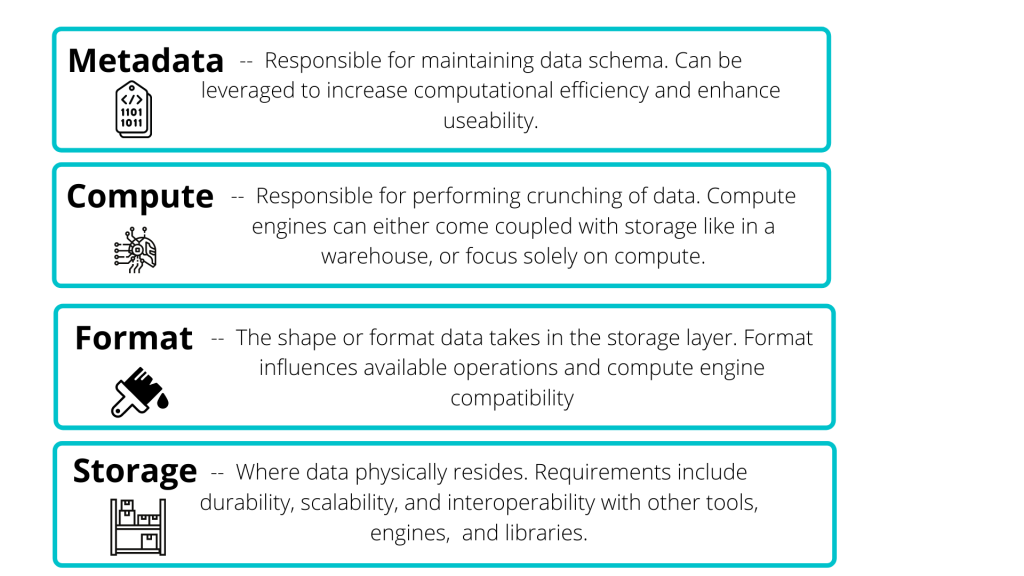
Data lakes are comprised of four primary components: storage, format, compute, and metadata layers [source].
A data lake is a centralized repository for large amounts of data from a variety of sources. Data can be structured, semi-structured, or unstructured in general. The goal is to rapidly ingest data and make it available to or accessible to other team members such as data scientists, analysts, and engineers. The data lake is widely used for machine learning and analytical solutions. Generally, when you store data in a data lake, you associate it with some form of metadata to facilitate access. Generally, a data lake solution must be secure and scalable. Additionally, the hardware should be affordable. The reason for this is that you want to store as much data as possible quickly.
Data Lake vs Data Warehouse
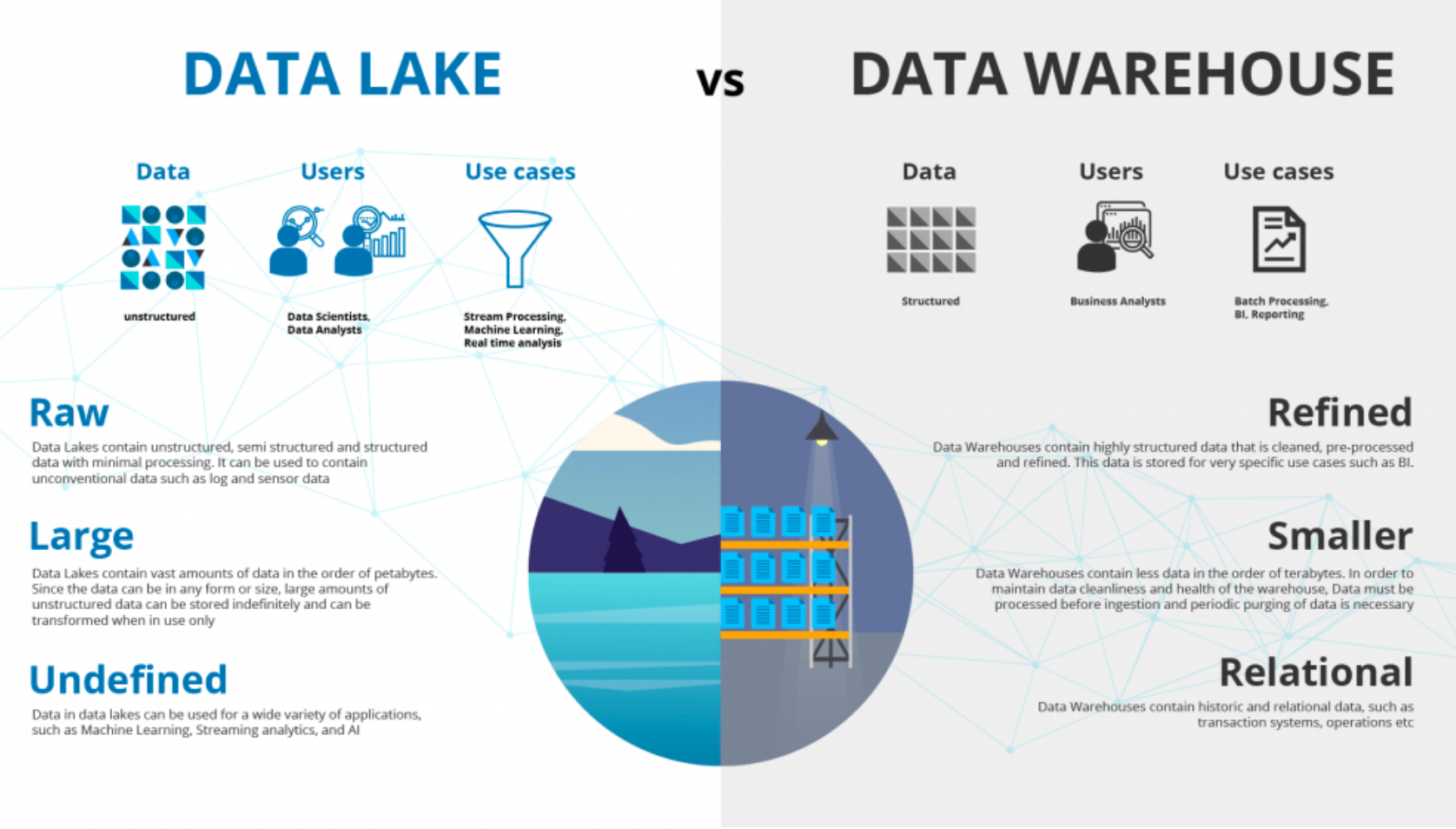
Generally a data lake is an unstructured data and the target users are data scientists or data analysts. It stores huge amount of data, sometimes in the size of petabytes and terabytes. The use cases which are covered by data lake are basically stream processing, machine learning, and real-time analytics. On the data warehouse side, the data is generally structured. The users are business analysts, the data size is generally small, and the use case consists of batch processing or BI reporting.
ETL vs ELT
- Extract Transform and Load vs Extract Load and Transform
- ETL is mainly used for a small amount of data whereas ELT is used for large amounts of data
- ELT provides data lake support (Schema on read)
- ETL provides data warehouse solutions
Data lake solutions provided by main cloud providers are as follows:
- GCP - cloud storage
- AWS - S3
- AZURE - AZURE BLOB
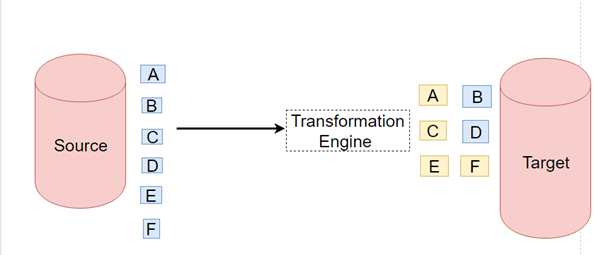
We saw a simple data pipeline in week 1. One of the problems in that data pipeline was that we did several important jobs in the same place: downloading data and doing small processing and putting it into postgres. What if after downloading data, some error happens in the code or with the internet? We will lose the downloaded data and should do everything from scratch. That's why we need to do those steps separately.
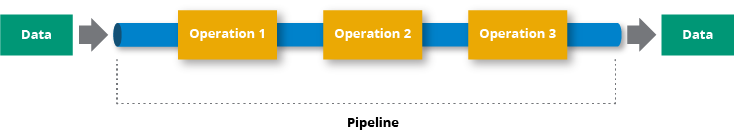
A data pipeline is a series of steps for data processing. If the data has not yet been loaded into the data platform, it is ingested at the pipeline's start. Then there is a series of steps, each of which produces an output that serves as the input for the subsequent step. This procedure is repeated until the pipeline is completed. In some instances, independent steps may be performed concurrently. [source].
A data pipeline is composed of three critical components: a source, a processing step or series of processing steps, and a destination. The destination may be referred to as a sink in some data pipelines. Data pipelines, for example, enable the flow of data from an application to a data warehouse, from a data lake to an analytics database, or to a payment processing system. Additionally, data pipelines can share the same source and sink, allowing the pipeline to focus entirely on data modification. When data is processed between points A and B (or B, C, and D), there is a data pipeline between those points [source].
In our example, the data pipeline we had in the previous week can be as follows:
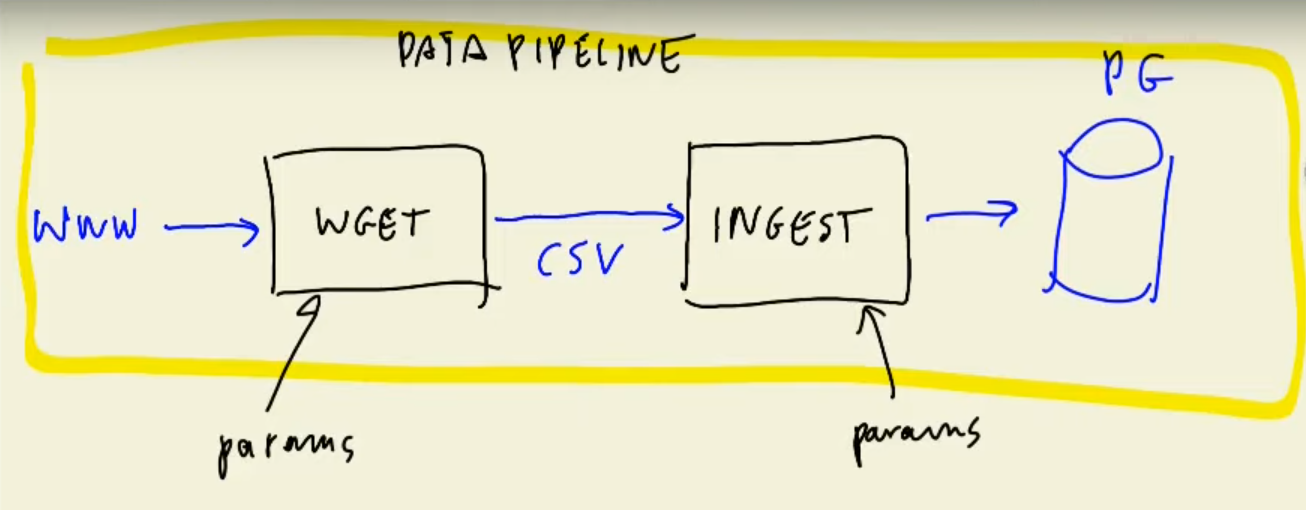
We separated downloading dataset using wget and then ingesting it into postgres. I think we can have even another more step for processing (changing the string to datetime in the downloaded dataset).
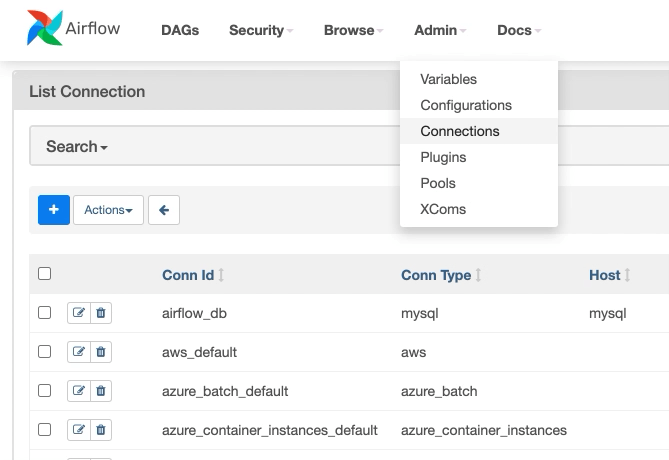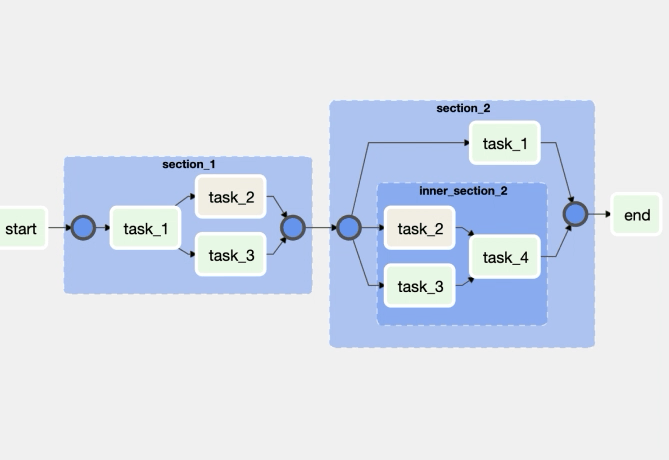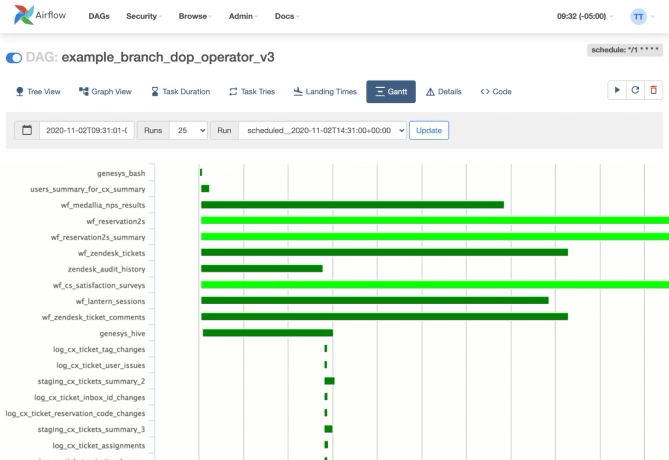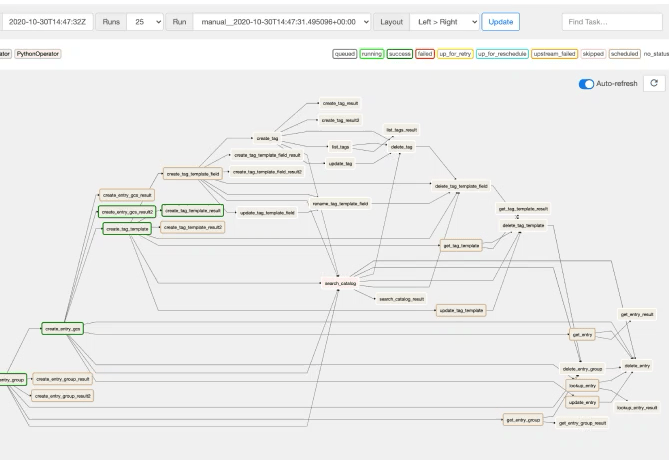
But this week, we will do something more complex. Let's have a look at the data workflow.
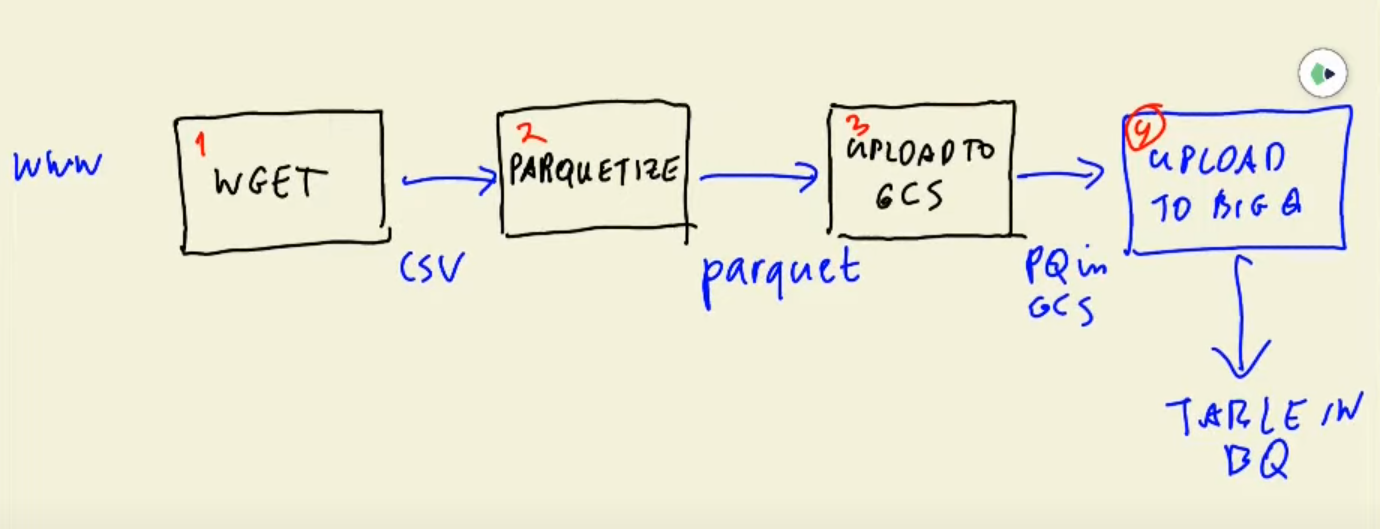
The above figure is called a DAG (Directed Acyclic Graph). We need to be sure that all steps are done sequentially and we can retry some of the steps if some thing happens and then go to the next step. There are some tools called workflow engines tat allow us to define these DAGs and do the data workflow orchestration:
- LUIGI
- APACHE AIRFLOW (we will go for this)
- PREFECT
- Google Cloud Dataflow
Let's get more familiar with the last two ones:
Airflow
Airflow is a platform to programmatically author, schedule and monitor workflows. Use Airflow to author workflows as Directed Acyclic Graphs (DAGs) of tasks. The Airflow scheduler executes your tasks on an array of workers while following the specified dependencies. Rich command line utilities make performing complex surgeries on DAGs a snap. The rich user interface makes it easy to visualize pipelines running in production, monitor progress, and troubleshoot issues when needed. When workflows are defined as code, they become more maintainable, versionable, testable, and collaborative [Airflow docs].
Google Cloud Dataflow
Real-time data is generated by websites, mobile applications, IoT devices, and other workloads. All businesses make data collection, processing, and analysis a priority. However, data from these systems is frequently not in a format suitable for analysis or effective use by downstream systems. That is where Dataflow enters the picture! Dataflow is used to process and enrich batch or stream data for analysis, machine learning, and data warehousing applications.
Dataflow is a serverless, high-performance, and cost-effective service for stream and batch processing. It enables portability for processing jobs written in the open source Apache Beam libraries and alleviates operational burden on your data engineering teams by automating infrastructure provisioning and cluster management [Google cloud docs].
Here is a comparison between Airflow and Google cloud dataflow.
Google Cloud Composer
Google also provides a fully managed service for workflow orchestration built on the Apache Airflow called Google Cloud Composer.
Cloud Composer is a fully managed workflow orchestration service, enabling you to create, schedule, monitor, and manage workflows that span across clouds and on-premises data centers. Cloud Composer is built on the popular Apache Airflow open source project and operates using the Python programming language. By using Cloud Composer instead of a local instance of Apache Airflow, you can benefit from the best of Airflow with no installation or management overhead. Cloud Composer helps you create Airflow environments quickly and use Airflow-native tools, such as the powerful Airflow web interface and command-line tools, so you can focus on your workflows and not your infrastructure [gcp docs].
Here is a good comparison between Google Cloud Composer and Airflow on Docker and on-premise:
Cloud Composer is a GCP managed service for Airflow. Composer runs in something known as a Composer environment, which runs on Google Kubernetes Engine cluster. It also makes use of various other GCP services such as:>> - Cloud SQL - stores the metadata associated with Airflow,
App Engine Flex - Airflow web server runs as an App Engine Flex application, which is protected using an Identity-Aware Proxy,
GCS bucket - in order to submit a pipeline to be scheduled and run on Composer, all that we need to do is to copy out Python code into a GCS bucket. Within that, it'll have a folder called DAGs. Any Python code uploaded into that folder is automatically going to be picked up and processed by Composer.
How Cloud Composer benefits?
Focus on your workflows, and let Composer manage the infrastructure (creating the workers, setting up the web server, the message brokers),
One-click to create a new Airflow environment,
Easy and controlled access to the Airflow Web UI,
Provide logging and monitoring metrics, and alert when your workflow is not running,
Integrate with all of Google Cloud services:Big Data, Machine Learning and so on. Run jobs elsewhere, i.e. other cloud provider (Amazon).> > Of course you have to pay for the hosting service, but the cost is low compare to if you have to host a production airflow server on your own.
Airflow on-premise
- DevOps work that need to be done:create a new server, manage Airflow installation, takes care of dependency and package management, check server health, scaling and security.>
pull an Airflow image from a registry and creating the container
creating a volume that maps the directory on local machine where DAGs are held, and the locations where Airflow reads them on the container,
whenever you want to submit a DAG that needs to access GCP service, you need to take care of setting up credentials. Application's service account should be created and downloaded as a JSON file that contains the credentials. This JSON file must be linked into your docker container and the GOOGLE_APPLICATION_CREDENTIALS environment variable must contain the path to the JSON file inside the container.
To sum up, if you don’t want to deal with all of those DevOps problem, and instead just want to focus on your workflow, then Google Cloud composer is a great solution for you.
To compare Google Dataflow and Google Cloud Composer, I found this stackoverflow answer very interesting:
For the basics of your described task, Cloud Dataflow is a good choice. Big data that can be processed in parallel is a good choice for Cloud Dataflow.
The real world of processing big data is usually messy. Data is usually somewhat to very dirty, arrives constantly or in big batches and needs to be processed in time sensitive ways. Usually it takes the coordination of more than one task / system to extract desired data. Think of load, transform, merge, extract and store types of tasks. Big data processing is often glued together using using shell scripts and / or Python programs. This makes automation, management, scheduling and control processes difficult.
Google Cloud Composer is a big step up from Cloud Dataflow. Cloud Composer is a cross platform orchestration tool that supports AWS, Azure and GCP (and more) with management, scheduling and processing abilities.
Cloud Dataflow handles tasks. Cloud Composer manages entire processes coordinating tasks that may involve BigQuery, Dataflow, Dataproc, Storage, on-premises, etc.
If you need / require more management, control, scheduling, etc. of your big data tasks, then Cloud Composer adds significant value. If you are just running a simple Cloud Dataflow task on demand once in a while, Cloud Composer might be overkill.
In this post, we just review Airflow and how to use it on Docker. Let's get started.
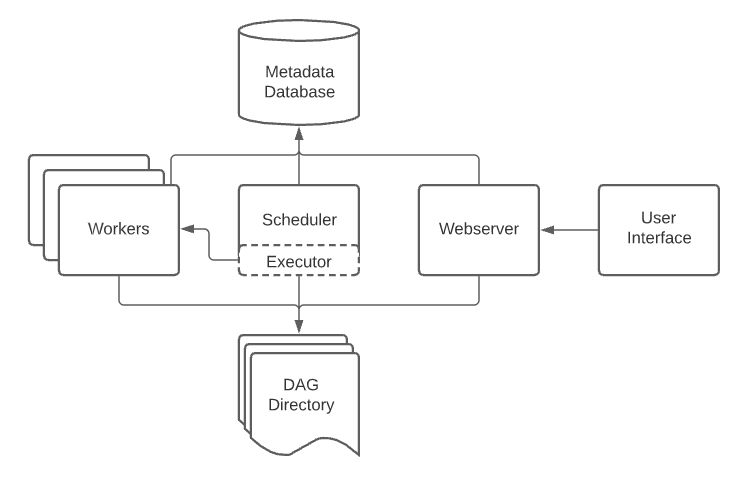
Let's review the Airflow architecture. An Airflow installation generally consists of the following components:
-
Web server: GUI to inspect, trigger and debug the behaviour of DAGs and tasks. Available at http://localhost:8080.
-
Scheduler: Responsible for scheduling jobs. Handles both triggering & scheduled workflows, submits Tasks to the executor to run, monitors all tasks and DAGs, and then triggers the task instances once their dependencies are complete.
-
Worker: This component executes the tasks given by the scheduler.
-
Metadata database (postgres): Backend to the Airflow environment. Used by the scheduler, executor and webserver to store state.
Other components (seen in docker-compose services):
- redis: Message broker that forwards messages from scheduler to worker.
- flower: The flower app for monitoring the environment. It is available at http://localhost:5555.
- airflow-init: initialization service (customized as per this design)
Please read more about Airflow architecture here before continuing the blog post.
Now let's install airflow environment using docker.
You may need Python version 3.7+.
You may also need to upgrade your docker-compose version to v2.x+ (as suggested in the course - however airflow documentation suggests v1.29.1 or newer).
Default amount of memory available for Docker on MacOS is often not enough to get Airflow up and running. If enough memory is not allocated, it might lead to airflow webserver continuously restarting. You should at least allocate 4GB memory for the Docker Engine (ideally 8GB). You can check and change the amount of memory in Resources
You can also check if you have enough memory by running this command [Airflow docs]:
docker run --rm "debian:buster-slim" bash -c 'numfmt --to iec $(echo $(($(getconf _PHYS_PAGES) * $(getconf PAGE_SIZE))))'
For me, this is 16 GB:
Unable to find image 'debian:buster-slim' locally
buster-slim: Pulling from library/debian
6552179c3509: Pull complete
Digest: sha256:f6e5cbc7eaaa232ae1db675d83eabfffdabeb9054515c15c2fb510da6bc618a7
Status: Downloaded newer image for debian:buster-slim
16G
If enough memory is not allocated, it might lead to airflow-webserver continuously restarting. I used this answer to update mine. For limiting memory, it is easy to do it in mac and windows like here and for linux you can check here.
To deploy Airflow on Docker Compose, you should fetch docker-compose.yaml.
curl -LfO 'https://airflow.apache.org/docs/apache-airflow/2.2.3/docker-compose.yaml'
This file contains several service definitions:
- airflow-scheduler - The scheduler monitors all tasks and DAGs, then triggers the task instances once their dependencies are complete. Behind the scenes, the scheduler spins up a subprocess, which monitors and stays in sync with all DAGs in the specified DAG directory. Once per minute, by default, the scheduler collects DAG parsing results and checks whether any active tasks can be triggered [ref].
- airflow-webserver - The webserver is available at http://localhost:8080.
- airflow-worker - The worker that executes the tasks given by the scheduler.
- airflow-init - The initialization service.
- flower - The flower app is a web based tool for monitoring the environment. It is available at http://localhost:5555.
- postgres - The database.
- redis - The redis - broker that forwards messages from scheduler to worker.
All these services allow you to run Airflow with CeleryExecutor. For more information, see Architecture Overview.
Some directories in the container are mounted, which means that their contents are synchronized between your computer and the container.
- ./dags - you can put your DAG files here.
- ./logs - contains logs from task execution and scheduler.
- ./plugins - you can put your custom plugins here. Airflow has a simple plugin manager built-in that can integrate external features to its core by simply dropping files in your $AIRFLOW_HOME/plugins folder.
Here is the architecture of docker-compose.yaml file:
version: '3'
x-airflow-common:
&airflow-common
# In order to add custom dependencies or upgrade provider packages you can use your extended image.
# Comment the image line, place your Dockerfile in the directory where you placed the docker-compose.yaml
# and uncomment the "build" line below, Then run `docker-compose build` to build the images.
image: ${AIRFLOW_IMAGE_NAME:-apache/airflow:2.2.3}
# build: .
environment:
&airflow-common-env
AIRFLOW__CORE__EXECUTOR: CeleryExecutor
AIRFLOW__CORE__SQL_ALCHEMY_CONN: postgresql+psycopg2://airflow:airflow@postgres/airflow
AIRFLOW__CELERY__RESULT_BACKEND: db+postgresql://airflow:airflow@postgres/airflow
AIRFLOW__CELERY__BROKER_URL: redis://:@redis:6379/0
AIRFLOW__CORE__FERNET_KEY: ''
AIRFLOW__CORE__DAGS_ARE_PAUSED_AT_CREATION: 'true'
AIRFLOW__CORE__LOAD_EXAMPLES: 'true'
AIRFLOW__API__AUTH_BACKEND: 'airflow.api.auth.backend.basic_auth'
_PIP_ADDITIONAL_REQUIREMENTS: ${_PIP_ADDITIONAL_REQUIREMENTS:-}
volumes:
- ./dags:/opt/airflow/dags
- ./logs:/opt/airflow/logs
- ./plugins:/opt/airflow/plugins
user: "${AIRFLOW_UID:-50000}:0"
depends_on:
&airflow-common-depends-on
redis:
condition: service_healthy
postgres:
condition: service_healthy
services:
postgres:
...
redis:
...
airflow-webserver:
...
airflow-scheduler:
...
airflow-worker:
...
airflow-triggerer:
...
airflow-init:
...
airflow-cli:
...
flower:
...
volumes:
postgres-db-volume:
The above file uses the latest Airflow image (apache/airflow). If you need to install a new Python library or system library, you can build your image.
When running Airflow locally, you may wish to use an extended image that includes some additional dependencies - for example, you may wish to add new Python packages or upgrade the airflow providers to a newer version. This is accomplished by including a custom Dockerfile alongside your docker-compose.yaml file. Then, using the docker-compose build command, you can create your image (you need to do it only once). Additionally, you can add the --build flag to your docker-compose commands to automatically rebuild the images when other docker-compose commands are run. To learn more and see additional examples, visit here [Airflow docs].
cd ~ && mkdir -p ~/.google/credentials/
mv <path/to/your/service-account-authkeys>.json ~/.google/credentials/google_credentials.json
In order to use airflow with GCP, we have changed the docker-compose.yaml file in this course as follows:
- instead of using the official airflow image as the base image, we use a custom docker file to build and start from.
build:
context: .
dockerfile: ./Dockerfile
- disable loading the DAG examples that ship with Airflow. It’s good to get started, but you probably want to set this to False in a production environment
AIRFLOW__CORE__LOAD_EXAMPLES: 'false'
- add GCP environment variables (you need to use your own gcp project id and the gcs bucket you created in the previous week)
GOOGLE_APPLICATION_CREDENTIALS: /.google/credentials/google_credentials.json
AIRFLOW_CONN_GOOGLE_CLOUD_DEFAULT: 'google-cloud-platform://?extra__google_cloud_platform__key_path=/.google/credentials/google_credentials.json'
GCP_PROJECT_ID: 'pivotal-surfer-336713'
GCP_GCS_BUCKET: "dtc_data_lake_pivotal-surfer-336713"
- add the folder we created at the beginning of the post for google credentials.
- ~/.google/credentials/:/.google/credentials:ro
Here is the beginning of the file after our modifications:
build:
context: .
dockerfile: ./Dockerfile
environment:
&airflow-common-env
AIRFLOW__CORE__EXECUTOR: CeleryExecutor
AIRFLOW__CORE__SQL_ALCHEMY_CONN: postgresql+psycopg2://airflow:airflow@postgres/airflow
AIRFLOW__CELERY__RESULT_BACKEND: db+postgresql://airflow:airflow@postgres/airflow
AIRFLOW__CELERY__BROKER_URL: redis://:@redis:6379/0
AIRFLOW__CORE__FERNET_KEY: ''
AIRFLOW__CORE__DAGS_ARE_PAUSED_AT_CREATION: 'true'
AIRFLOW__CORE__LOAD_EXAMPLES: 'false'
AIRFLOW__API__AUTH_BACKEND: 'airflow.api.auth.backend.basic_auth'
_PIP_ADDITIONAL_REQUIREMENTS: ${_PIP_ADDITIONAL_REQUIREMENTS:-}
GOOGLE_APPLICATION_CREDENTIALS: /.google/credentials/google_credentials.json
AIRFLOW_CONN_GOOGLE_CLOUD_DEFAULT: 'google-cloud-platform://?extra__google_cloud_platform__key_path=/.google/credentials/google_credentials.json'
GCP_PROJECT_ID: 'pivotal-surfer-336713'
GCP_GCS_BUCKET: "dtc_data_lake_pivotal-surfer-336713"
volumes:
- ./dags:/opt/airflow/dags
- ./logs:/opt/airflow/logs
- ./plugins:/opt/airflow/plugins
- ~/.google/credentials/:/.google/credentials:ro
Following is the custom Dockerfile whcich is placed inside the airflow folder.
The Dockerfile has the custom packages to be installed. The one we'll need the most is gcloud to connect with the GCS bucket/Data Lake.
# First-time build can take upto 10 mins.
FROM apache/airflow:2.2.3
ENV AIRFLOW_HOME=/opt/airflow
USER root
RUN apt-get update -qq && apt-get install vim -qqq
# git gcc g++ -qqq
COPY requirements.txt .
RUN pip install --no-cache-dir -r requirements.txt
# Ref: https://airflow.apache.org/docs/docker-stack/recipes.html
SHELL ["/bin/bash", "-o", "pipefail", "-e", "-u", "-x", "-c"]
ARG CLOUD_SDK_VERSION=322.0.0
ENV GCLOUD_HOME=/home/google-cloud-sdk
ENV PATH="${GCLOUD_HOME}/bin/:${PATH}"
RUN DOWNLOAD_URL="https://dl.google.com/dl/cloudsdk/channels/rapid/downloads/google-cloud-sdk-${CLOUD_SDK_VERSION}-linux-x86_64.tar.gz" \
&& TMP_DIR="$(mktemp -d)" \
&& curl -fL "${DOWNLOAD_URL}" --output "${TMP_DIR}/google-cloud-sdk.tar.gz" \
&& mkdir -p "${GCLOUD_HOME}" \
&& tar xzf "${TMP_DIR}/google-cloud-sdk.tar.gz" -C "${GCLOUD_HOME}" --strip-components=1 \
&& "${GCLOUD_HOME}/install.sh" \
--bash-completion=false \
--path-update=false \
--usage-reporting=false \
--quiet \
&& rm -rf "${TMP_DIR}" \
&& gcloud --version
WORKDIR $AIRFLOW_HOME
USER $AIRFLOW_UID
The requirements.txt file in the Dockerfile which contains the required pyton packages is as follows:
apache-airflow-providers-google
pyarrow
In case you don't want to see so many services as it is done in the above docker-compose.yaml file, you can use the following one which is placed in the week_2_data_ingestion/airflow/extras folder in the course github repo:
version: '3.7'
services:
webserver:
container_name: airflow
build:
context: ..
dockerfile: ../Dockerfile
environment:
- PYTHONPATH=/home/airflow
# airflow connection with SQLAlchemy container
- AIRFLOW__CORE__SQL_ALCHEMY_CONN=sqlite:///$AIRFLOW_HOME/airflow.db
- AIRFLOW__CORE__EXECUTOR=LocalExecutor
# disable example loading
- AIRFLOW__CORE__LOAD_EXAMPLES=FALSE
volumes:
- ./dags:/home/airflow/dags
# user: "${AIRFLOW_UID:-50000}:0"
ports:
- "8080:8080"
command: > # airflow db upgrade;
bash -c "
airflow scheduler -D;
rm /home/airflow/airflow-scheduler.*;
airflow webserver"
healthcheck:
test: [ "CMD-SHELL", "[ -f /home/airflow/airflow-webserver.pid ]" ]
interval: 30s
timeout: 30s
retries: 3
We will not use this file to avoid any confusion.
There is also another lightweight and less memory-intensive docker-compose file in the github repo which can be used.
There is another one from one of the students here which sounds interesting too.
Before starting Airflow for the first time, You need to prepare your environment, i.e. create the necessary files, directories and initialize the database.
On Linux, the quick-start needs to know your host user id and needs to have group id set to 0. Otherwise the files created in dags, logs and plugins will be created with root user. You have to make sure to configure them for the docker-compose: (run it inside the airflow folder where the docker-compose.yaml file is placed) [Airflow docs]
mkdir -p ./dags ./logs ./plugins
echo -e "AIRFLOW_UID=$(id -u)" > .env
For other operating systems, you will get warning that AIRFLOW_UID is not set, but you can ignore it. You can also manually create the .env file in the same folder your docker-compose.yaml is placed with this content to get rid of the warning:
AIRFLOW_UID=1000
Read more about environment variables for compose here. It seems that when we run docker-compose up, it looks for .env file in the same directory and uses the variables in that file.
Then we need to initialize the database. On all operating systems, you need to run database migrations and create the first user account. To do it, run.
docker-compose build
docker-compose up airflow-init
docker-compose up
You may also some error, but you can ignore them as they are for some services in the official docker compose file that we do not use.
You can check which services are up using:
docker-compose ps
For me, the output is as follows:
airflow-airflow-scheduler-1 "/usr/bin/dumb-init …" airflow-scheduler running (healthy) 8080/tcp
airflow-airflow-triggerer-1 "/usr/bin/dumb-init …" airflow-triggerer running (healthy) 8080/tcp
airflow-airflow-webserver-1 "/usr/bin/dumb-init …" airflow-webserver running (healthy) 0.0.0.0:8080->8080/tcp, :::8080->8080/tcp
airflow-airflow-worker-1 "/usr/bin/dumb-init …" airflow-worker running (healthy) 8080/tcp
airflow-flower-1 "/usr/bin/dumb-init …" flower running (healthy) 0.0.0.0:5555->5555/tcp, :::5555->5555/tcp
airflow-postgres-1 "docker-entrypoint.s…" postgres running (healthy) 5432/tcp
airflow-redis-1 "docker-entrypoint.s…" redis running (healthy) 6379/tcp
There are several ways to interact with it:
- by running CLI commands.
- via a browser using the web interface.
- using the REST API.
For the web interface, you can go to this address: http://0.0.0.0:8080/
The airflow UI will be like this:
The account created has the login airflow and the password airflow. After log in you will see two generated dags from the week_2_data_ingestion/airflow/dags folder.
A Workflow has the following components:
-
DAG: Directed acyclic graph, specifies the dependencies between a set of tasks with explicit execution order, and has a beginning as well as an end. (Hence, “acyclic”)-
DAG Structure: DAG Definition, Tasks (eg. Operators), Task Dependencies (control flow:>>or<<)
-
-
Task: a defined unit of work (aka, operators in Airflow). The Tasks themselves describe what to do, be it fetching data, running analysis, triggering other systems, or more.- Common Types: Operators (used in this workshop), Sensors, TaskFlow decorators
- Sub-classes of Airflow's BaseOperator
-
DAG Run: individual execution/run of a DAG- scheduled or triggered
-
Task Instance: an individual run of a single task. Task instances also have an indicative state, which could be “running”, “success”, “failed”, “skipped”, “up for retry”, etc.- Ideally, a task should flow from
none, toscheduled, toqueued, torunning, and finally tosuccess.
- Ideally, a task should flow from
Let's look at how to use Airflow to ingest data into GCP. To do so, we'll need to create a DAG object. One thing to remember is that this Airflow Python script is really just a configuration file that specifies the DAG's structure as code. The tasks defined here will be executed in a context distinct from that of this script. This script cannot be used to cross-communicate between tasks because different tasks run on different workers at different times. Note that we have a more advanced feature called XComs that can be used for this purpose [Airflow docs].
People mistakenly believe that the DAG definition file is where they can do actual data processing - this is not the case! The goal of the script is to create a DAG object. It must evaluate quickly (seconds, not minutes) because the scheduler will run it on a regular basis to reflect any changes [Airflow docs].
The structure of a DAG file is as follows:
# Imports
from airflow import DAG
...
default_args = {
...
}
with DAG(
'tutorial',
default_args=default_args,
description='A simple tutorial DAG',
schedule_interval=timedelta(days=1),
start_date=datetime(2021, 1, 1),
catchup=False,
tags=['example'],
) as dag:
# t1, t2 and t3 are examples of tasks created by instantiating operators
t1 = BashOperator(
task_id='print_date',
bash_command='date',
)
t2 = ...
t3 = ...
## defining task dependencies
t1 >> [t2, t3]
- An Airflow pipeline is just a Python script that happens to define an Airflow DAG object.
- We have the choice to explicitly pass a set of arguments to each task’s constructor (which would become redundant), or (better!) we can define a dictionary of default parameters that we can use when creating tasks.
- We’ll need a DAG object to nest our tasks into. Here we pass a string that defines the dag_id, which serves as a unique identifier for your DAG. We also pass the default argument dictionary that we just defined and define a schedule_interval of 1 day for the DAG.
- Tasks are generated when instantiating operator objects. An object instantiated from an operator is called a task. The first argument task_id acts as a unique identifier for the task.
- Then we need to define dependencies between tasks.
You can check more tutorials and examples here.
Now let's see out own DAG file for ingesting yellow_tripdata_2021-01.csv dataset into GCP which is place in week_2_data_ingestion/airflow/dags/data_ingestion_gcs_dag.py. First let's check the structure:
- importing python libraries
- in-built airflow like
BashOperatorandPythonOperator. There is alsoDockerOperatorto run docker in docker! -
storagefrom google cloud library to interact with GCS. - BigQuery from google airflow provider to interact with BigQuery and create an external table.
-
pyarrowlibrary for converting dataset type toparquetbefore uploading it to GCS.parquetis used more in production and is faster to upload it and also uses less space on GCS.
- in-built airflow like
- setting some variables
-
GCP_PROJECT_ID,GCP_GCS_BUCKETwhich we set in thedocker-compose.yamlfile underx-airflow-commonsection. - info about dataset url
- airflow local folder path
- name of the desired parquet file
- BigQuery dataset which can be found from
variables.tffrom terraform folder of week 1 (inweek_1_basics_n_setup/1_terraform_gcp/terraform/variables.tf). I think the name wasBQ_DATASETthere, but the value is the sametrips_data_all.
-
- Some python functions which will be attached to
PythonOperators likeformat_to_parquet()andupload_to_gcs(). Their names describe their functionality. - Default arguments which will be used in DAG definition.
- Then the DAG declaration with tasks and their dependencies
-
download_dataset_taskto download the dataset using a bash command. -
format_to_parquet_taskwhich call theformat_to_parquet()function. -
local_to_gcs_taskwhich call theupload_to_gcs()function. -
bigquery_external_table_taskto extract schema and create a BigQuery table form the file uploaded to GCS. You can easily run SQL queries on this table.
-
- Then the workflow for direction of tasks:
download_dataset_task >> format_to_parquet_task >> local_to_gcs_task >> bigquery_external_table_task
import os
import logging
from airflow import DAG
from airflow.utils.dates import days_ago
from airflow.operators.bash import BashOperator
from airflow.operators.python import PythonOperator
from google.cloud import storage
from airflow.providers.google.cloud.operators.bigquery import BigQueryCreateExternalTableOperator
import pyarrow.csv as pv
import pyarrow.parquet as pq
PROJECT_ID = os.environ.get("GCP_PROJECT_ID")
BUCKET = os.environ.get("GCP_GCS_BUCKET")
dataset_file = "yellow_tripdata_2021-01.csv"
dataset_url = f"https://s3.amazonaws.com/nyc-tlc/trip+data/{dataset_file}"
path_to_local_home = os.environ.get("AIRFLOW_HOME", "/opt/airflow/")
parquet_file = dataset_file.replace('.csv', '.parquet')
BIGQUERY_DATASET = os.environ.get("BIGQUERY_DATASET", 'trips_data_all')
def format_to_parquet(src_file):
if not src_file.endswith('.csv'):
logging.error("Can only accept source files in CSV format, for the moment")
return
table = pv.read_csv(src_file)
pq.write_table(table, src_file.replace('.csv', '.parquet'))
# NOTE: takes 20 mins, at an upload speed of 800kbps. Faster if your internet has a better upload speed
def upload_to_gcs(bucket, object_name, local_file):
"""
Ref: https://cloud.google.com/storage/docs/uploading-objects#storage-upload-object-python
:param bucket: GCS bucket name
:param object_name: target path & file-name
:param local_file: source path & file-name
:return:
"""
# WORKAROUND to prevent timeout for files > 6 MB on 800 kbps upload speed.
# (Ref: https://github.com/googleapis/python-storage/issues/74)
storage.blob._MAX_MULTIPART_SIZE = 5 * 1024 * 1024 # 5 MB
storage.blob._DEFAULT_CHUNKSIZE = 5 * 1024 * 1024 # 5 MB
# End of Workaround
client = storage.Client()
bucket = client.bucket(bucket)
blob = bucket.blob(object_name)
blob.upload_from_filename(local_file)
default_args = {
"owner": "airflow",
"start_date": days_ago(1),
"depends_on_past": False,
"retries": 1,
}
# NOTE: DAG declaration - using a Context Manager (an implicit way)
with DAG(
dag_id="data_ingestion_gcs_dag",
schedule_interval="@daily",
default_args=default_args,
catchup=False,
max_active_runs=1,
tags=['dtc-de'],
) as dag:
download_dataset_task = BashOperator(
task_id="download_dataset_task",
bash_command=f"curl -sS {dataset_url} > {path_to_local_home}/{dataset_file}"
)
format_to_parquet_task = PythonOperator(
task_id="format_to_parquet_task",
python_callable=format_to_parquet,
op_kwargs={
"src_file": f"{path_to_local_home}/{dataset_file}",
},
)
# TODO: Homework - research and try XCOM to communicate output values between 2 tasks/operators
local_to_gcs_task = PythonOperator(
task_id="local_to_gcs_task",
python_callable=upload_to_gcs,
op_kwargs={
"bucket": BUCKET,
"object_name": f"raw/{parquet_file}",
"local_file": f"{path_to_local_home}/{parquet_file}",
},
)
bigquery_external_table_task = BigQueryCreateExternalTableOperator(
task_id="bigquery_external_table_task",
table_resource={
"tableReference": {
"projectId": PROJECT_ID,
"datasetId": BIGQUERY_DATASET,
"tableId": "external_table",
},
"externalDataConfiguration": {
"sourceFormat": "PARQUET",
"sourceUris": [f"gs://{BUCKET}/raw/{parquet_file}"],
},
},
)
download_dataset_task >> format_to_parquet_task >> local_to_gcs_task >> bigquery_external_table_task
Let's run it. First go to localhost:8080 and use airflow and airflow as username and password to log in. Then switch on the data_ingestion_gcs_dag and click on that to open and be able to see the tree. You can also swith to graph using the toolbar on top of the page.
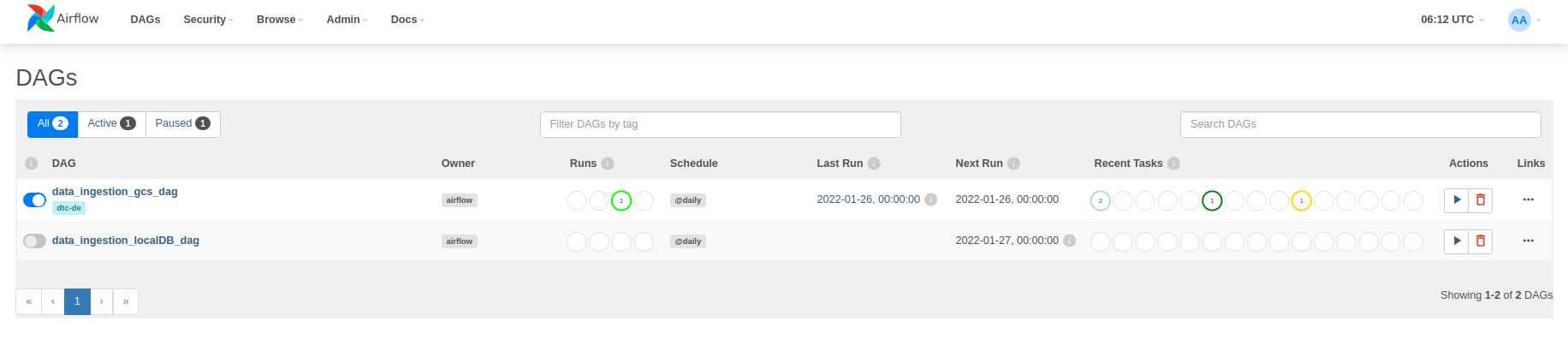
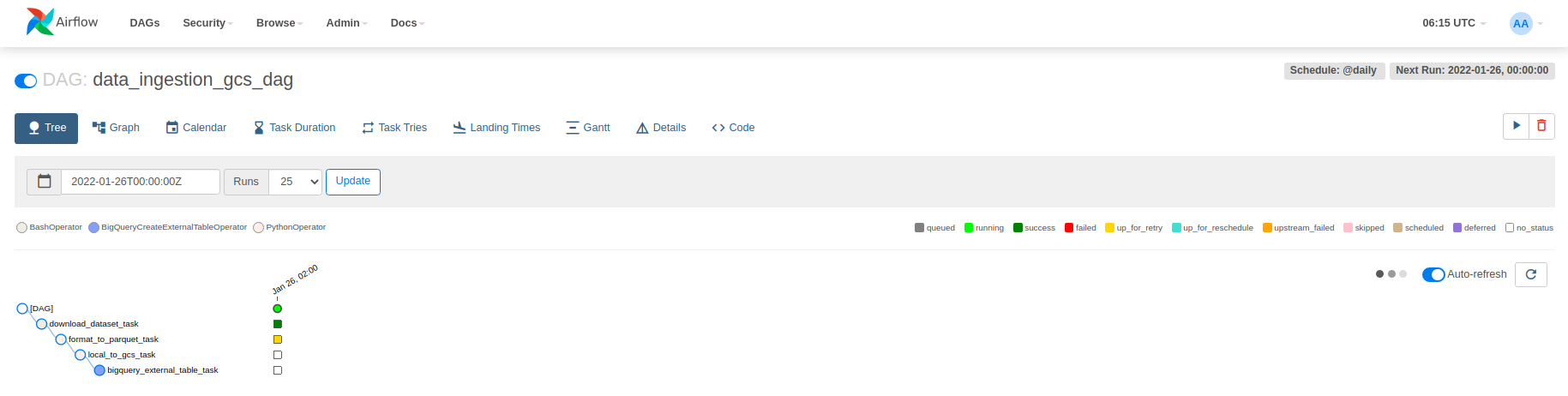
Then click on the play button on top-right part of the page and select Trigger DAG. Note that after running docker-compose ps everything should be in the healthy mode.
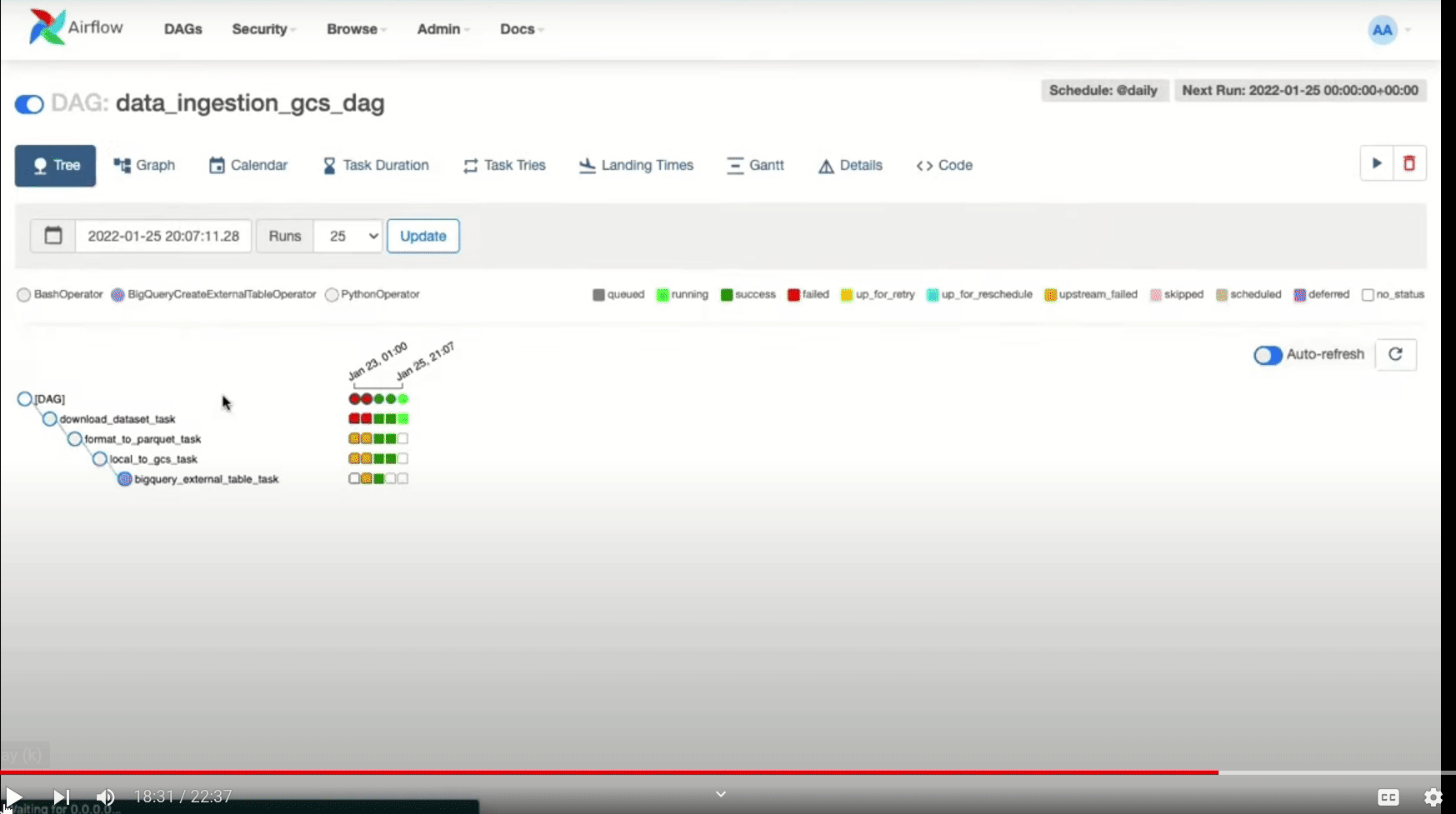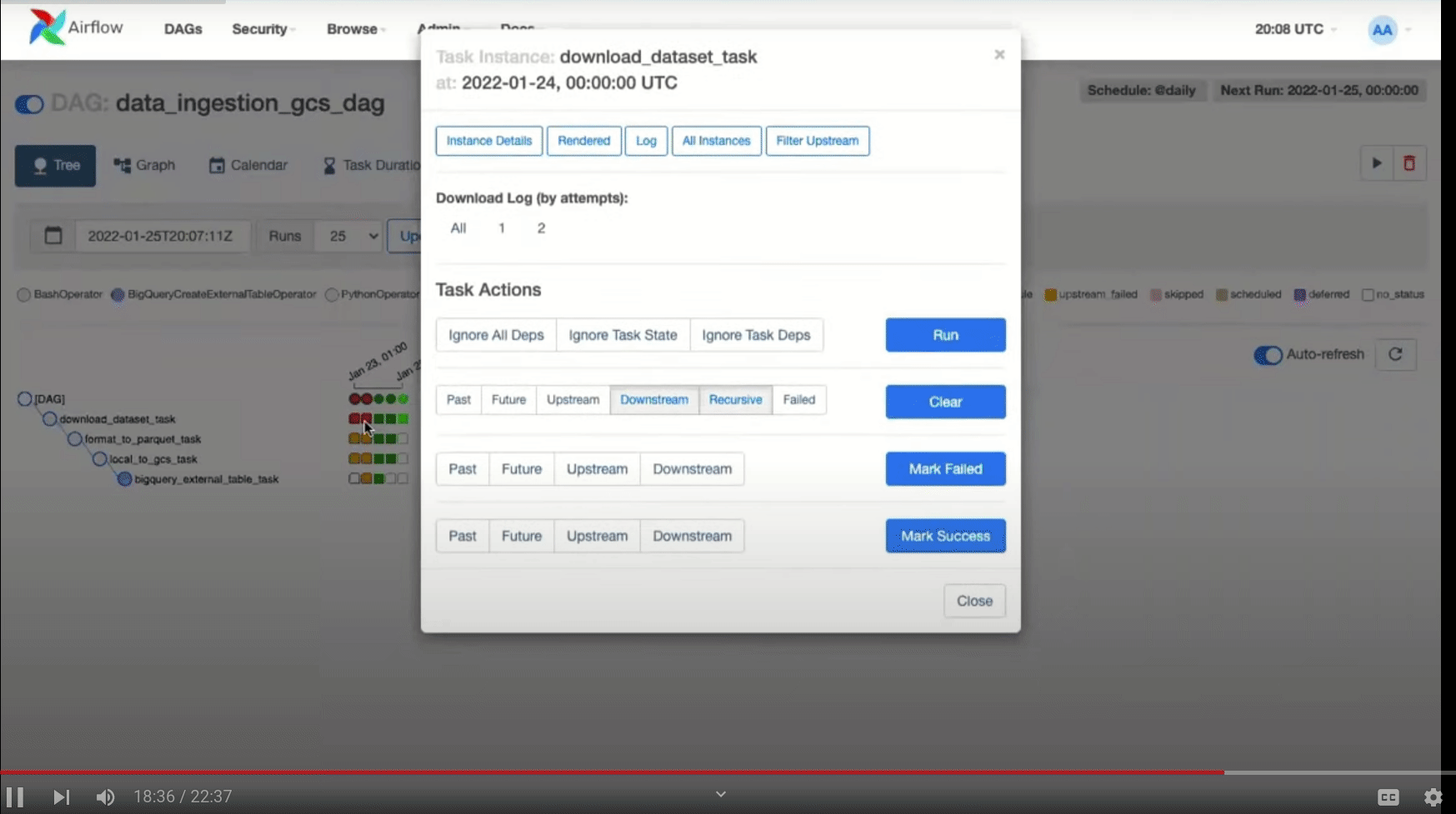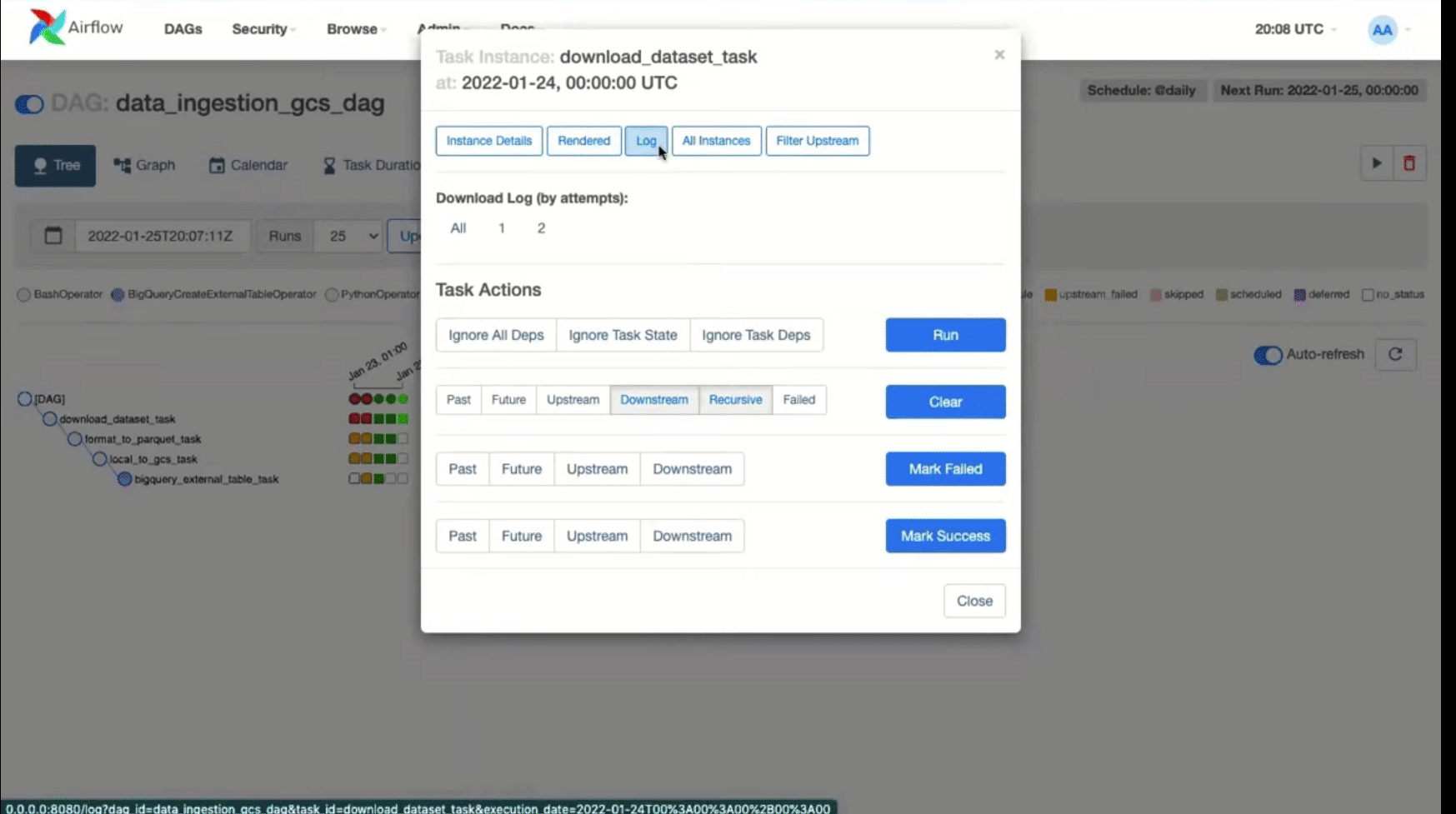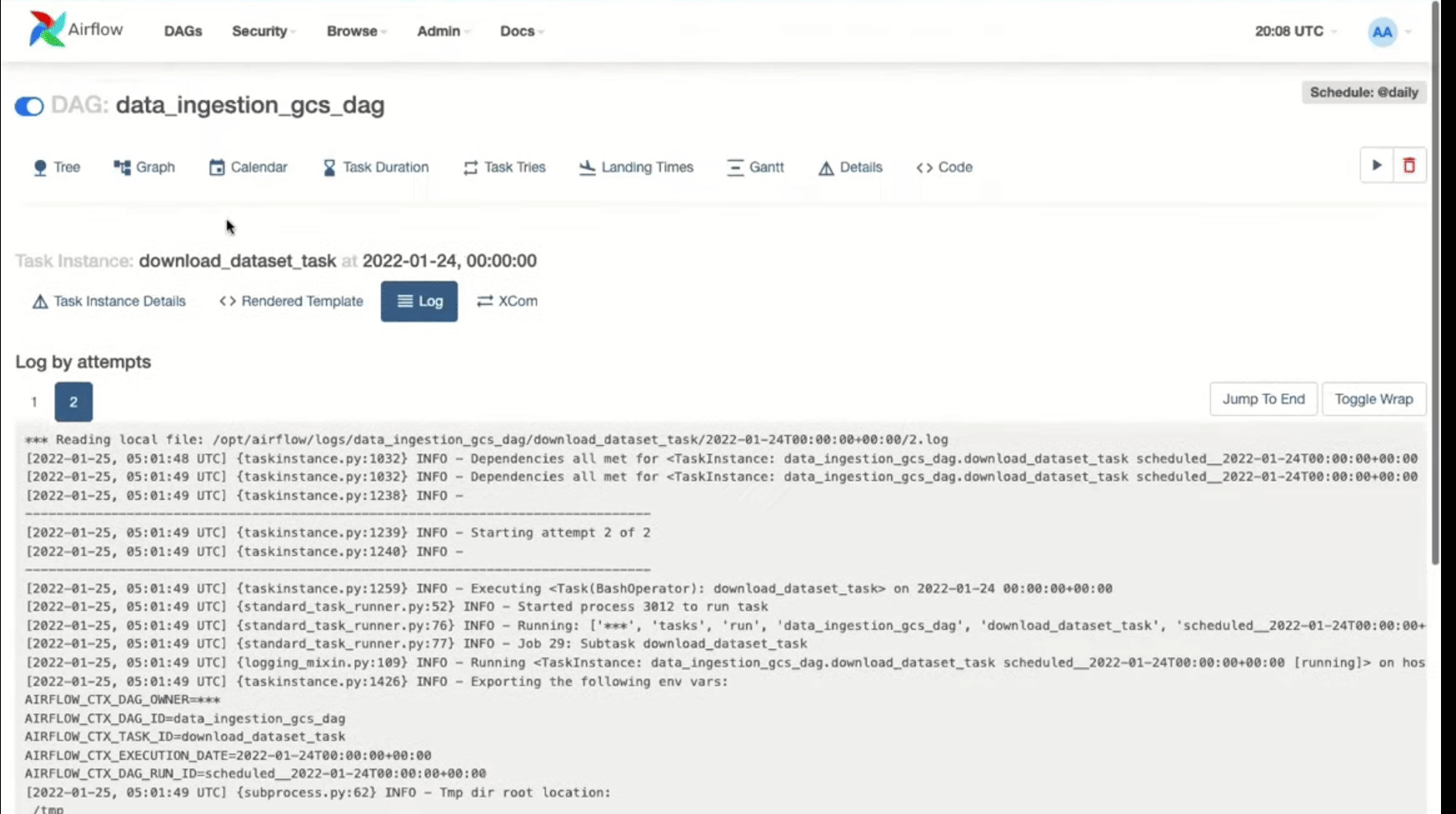
In case any of the tasks fails, you can check the logs as follows:
If the tasks are completed successfully, then you will see the uploaded parquet file in GCS bucket and also the table in BigQuery.
Note: All the PythonOperator codes (functions called by that) are executed in airflow-workers (which is a container) and files (datasets) are saved there and not in your local machine. If you use DockerOperator, you are actually running a docker inside another docker (airflow-worker).
On finishing your run or to shut down the container/s:
docker-compose down
To stop and delete containers, delete volumes with database data, and download images, run:
docker-compose down --volumes --rmi all
or
docker-compose down --volumes --remove-orphans
Before, we uploaded data to GCS using the upload_to_gcs function. But here, a s we want to ingest data into postgres (which we will run using another docker-compose), we need to use another function. All the steps are the same (check the above video) and we just check the DAG file and ingestion script here which is a modified version of what we used in week 1.
Let's see the week_2_data_ingestion/airflow/dags_local/data_ingestion_local.py file first:
import os
from datetime import datetime
from airflow import DAG
from airflow.operators.bash import BashOperator
from airflow.operators.python import PythonOperator
from ingest_script import ingest_callable
AIRFLOW_HOME = os.environ.get("AIRFLOW_HOME", "/opt/airflow/")
PG_HOST = os.getenv('PG_HOST')
PG_USER = os.getenv('PG_USER')
PG_PASSWORD = os.getenv('PG_PASSWORD')
PG_PORT = os.getenv('PG_PORT')
PG_DATABASE = os.getenv('PG_DATABASE')
local_workflow = DAG(
"LocalIngestionDag",
schedule_interval="0 6 2 * *",
start_date=datetime(2021, 1, 1)
)
URL_PREFIX = 'https://s3.amazonaws.com/nyc-tlc/trip+data'
URL_TEMPLATE = URL_PREFIX + '/yellow_tripdata_.csv'
OUTPUT_FILE_TEMPLATE = AIRFLOW_HOME + '/output_.csv'
TABLE_NAME_TEMPLATE = 'yellow_taxi_'
with local_workflow:
wget_task = BashOperator(
task_id='wget',
bash_command=f'curl -sSL {URL_TEMPLATE} > {OUTPUT_FILE_TEMPLATE}'
)
ingest_task = PythonOperator(
task_id="ingest",
python_callable=ingest_callable,
op_kwargs=dict(
user=PG_USER,
password=PG_PASSWORD,
host=PG_HOST,
port=PG_PORT,
db=PG_DATABASE,
table_name=TABLE_NAME_TEMPLATE,
csv_file=OUTPUT_FILE_TEMPLATE
),
)
wget_task >> ingest_task
And the ingestion function which is imported in the above script is as follows:
import os
from time import time
import pandas as pd
from sqlalchemy import create_engine
def ingest_callable(user, password, host, port, db, table_name, csv_file, execution_date):
print(table_name, csv_file, execution_date)
engine = create_engine(f'postgresql://{user}:{password}@{host}:{port}/{db}')
engine.connect()
print('connection established successfully, instering data...')
t_start = time()
df_iter = pd.read_csv(csv_file, iterator=True, chunksize=100000)
df = next(df_iter)
df.tpep_pickup_datetime = pd.to_datetime(df.tpep_pickup_datetime)
df.tpep_dropoff_datetime = pd.to_datetime(df.tpep_dropoff_datetime)
df.head(n=0).to_sql(name=table_name, con=engine, if_exists='replace')
df.to_sql(name=table_name, con=engine, if_exists='append')
t_end = time()
print('inserted the first chunk, took %.3f second' % (t_end - t_start))
while True:
t_start = time()
try:
df = next(df_iter)
except StopIteration:
print("completed")
break
df.tpep_pickup_datetime = pd.to_datetime(df.tpep_pickup_datetime)
df.tpep_dropoff_datetime = pd.to_datetime(df.tpep_dropoff_datetime)
df.to_sql(name=table_name, con=engine, if_exists='append')
t_end = time()
print('inserted another chunk, took %.3f second' % (t_end - t_start))
Now we have two docker-compose.yaml files: one for this week which runs airflow stuff and one for week 1 for postgres and pgadmin. Let's see how we can connect them.
When we run the docker-compose.yaml file for airflow, it creates a network called airflow_default which we will use as an external network to connect the docker-compose for postgres and pgadmin to [ref].
# docker-compose.yaml of week 1
services:
pgdatabase:
image: postgres:13
environment:
- POSTGRES_USER=root
- POSTGRES_PASSWORD=root
- POSTGRES_DB=ny_taxi
volumes:
- "./ny_taxi_postgres_data:/var/lib/postgresql/data:rw"
ports:
- "5432:5432"
networks:
- airflow
pgadmin:
image: dpage/pgadmin4
environment:
- PGADMIN_DEFAULT_EMAIL=admin@admin.com
- PGADMIN_DEFAULT_PASSWORD=root
ports:
- "8080:80"
networks:
airflow:
external:
name: airflow_default
We will have two postgres databases, one for airflow that stores its own metadata and one for uplading our dataset into.
First run docker-compose up in week 1 folder, and then test if you can connect using pgcli -h localhost -p 5432 -U root -d ny_taxi command.
Then we can go to the airflow worker container and see if we can connect to the postgres database that we ran above:
docker exec -it <container id> bash
# then type python to open python
> > from sqlalchemy import create_engine
>> engine = create_engine('postgresql://root:root@pgdatabase:5432/ny_taxi')>> engine.connect()
# no error here
If you don't see any error by following the above procedure, it shows that you can connect to the postgres from airflow worker container. So calling the ingestion script from the PythonOperator in the DAG file should work. Now you can run both docker-compose files and go to airflow web interface to see the DAGs running.
On finishing your run or to shut down the container/s:
docker-compose down
To stop and delete containers, delete volumes with database data, and download images, run:
docker-compose down --volumes --rmi all
or
docker-compose down --volumes --remove-orphans
Transfer Service in GCP
Until now we have used airflow DAGs to download dataset and then push it into GCS. We can also use a service from GCP called transfer service to do this task directly from other cloud providers or local storages. Storage transfer service is a secure, low-cost services for transferring data from cloud, like AWS or Azure, or on-premises sources. If you search for the transfer service in GCP, there are two separate services, one for cloud and one for on-premises.
To activate a job, you can use terraform or the UI in GCP. Check the following video to learn how to do it via GCP.
To learn how to do it via terraform, please check the following video:
Conclusion
The reviewed procedure in this post for Airflow is not really suitable for production. Usually a combination of Airflow & Kubernetes & Git can be used. You can check the following resources to learn more.
Deploying Apache Airflow to Google Kubernetes Engine
You can also use the Google Cloud Composer service to have a fully managed Airflow. This would be easier with a bit more cost.
A Cloud Composer environment is a wrapper around Apache Airflow. Cloud Composer creates the following components for each environment: [gcp docs]
- GKE cluster: The Airflow schedulers, workers, and Redis Queue run as GKE workloads on a single cluster, and are responsible for processing and executing DAGs. The cluster also hosts other Cloud Composer components like Composer Agent and Airflow Monitoring, which help manage the Cloud Composer environment, gather logs to store in Cloud Logging, and gather metrics to upload to Cloud Monitoring.
- Web server: The web server runs the Apache Airflow web interface, and Identity-Aware Proxy protects the interface. For more information, see Airflow Web Interface.
- Database: The database holds the Apache Airflow metadata.
- Cloud Storage bucket: Cloud Composer associates a Cloud Storage bucket with the environment. The associated bucket stores the DAGs, logs, custom plugins, and data for the environment. For more information about the storage bucket for Cloud Composer, see Data Stored in Cloud Storage.
To access and manage your Airflow environments, you can use the following Airflow-native tools:
- Web interface: You can access the Airflow web interface from the Google Cloud Console or by direct URL with the appropriate permissions. For information, see Airflow Web Interface.
- Command line tools: After you install the Google Cloud CLI, you can run gcloud composer environments commands to issue Airflow command-line commands to Cloud Composer environments. For information, see [Airflow Command-line Interface][cc-access-airflow-cli].
In addition to native tools, the Cloud Composer REST and RPC APIs provide programmatic access to your Airflow environments.